Like the fingerprints that came into use by detectives and police labs during the 1930s, each person has a unique DNA fingerprint. Unlike a conventional fingerprint that occurs only on the fingertips and can be altered by surgery, a DNA fingerprint is the same for every cell, tissue, and organ of a person. It cannot be altered by any known treatment. Consequently, DNA fingerprinting is rapidly becoming the primary method for identifying and distinguishing among individual human beings. DNA fingerprints are also useful in several applications of human health care research and for diagnosis of inherited disorders. DNA fingerprinting is often used in the justice system to link suspects to biological evidence and solve paternity cases.
How is DNA fingerprinting used to identify a criminal?
Oxford Education (YouTube)
The Polymerase Chain Reaction
PCR Set the Stage for a Scientific Revolution
In 1983, Kary Mullis at Cetus Corporation developed a new technique, called the polymerase chain reaction (PCR). The objective of PCR is to produce a large amount of DNA in a test tube (in vitro), starting from only a trace amount. Technically speaking, this means the controlled replication of a DNA sequence, or gene, of interest. The template strands can be any form of double-stranded DNA such as genomic DNA. A researcher can take trace amounts of genomic DNA from a drop of blood, a single hair follicle, or a cheek cell (in theory, only a single template strand is needed to copy and generate millions of new identical DNA molecules) and make enough to study. Prior to PCR, this would have been impossible.
PCR Amplification
It is estimated that there are 30,00050,000 individual genes in the human genome. The true power of PCR is the ability to target and make millions of copies of (or amplify) a specific piece of DNA (or gene) out of a complete genome.
The recipe for a PCR amplification of DNA contains a simple mixture of ingredients which should be recognizable as the peices used to replicate DNA in the body:
- DNA template containing the intact sequence of DNA to be amplified
- Individual deoxynucleotides (A, T, G, and C) raw material of DNA
- DNA polymerase an enzyme that assembles the nucleotides into a new DNA chain
- Magnesium ions a cofactor (catalyst) required by DNA polymerase to create the DNA chain
- Oligonucleotide primers pieces of DNA complementary to the template that tell DNA polymerase exactly where to start making copies
- Salt buffer provides the optimum ionic environment and pH for the PCR reaction
The template DNA can be from blood, skin, semen or hair etc. When all the other components are combined under the right conditions, a copy of the original double-stranded template DNA molecule is made doubling the number of template strands. Each time this cycle is repeated, copies are made from copies and the number of template strands doubles from 2 to 4 to 8 to 16 and so on until after 20 cycles there are 1,048,554 exact copies of the target sequence.
PCR makes use of the same basic processes that cells use to duplicate their DNA.
- Complementary DNA strand hybridization
- DNA strand synthesis via DNA polymerase
The DNA primers are designed to flank a DNA sequence within the genome and thus provide the exact start signal for the DNA polymerase to bind and begin synthesizing (replicating) copies of that target DNA. Complementary strand hybridization takes place when different primers anneal, or bind to each of their respective complementary base sequences on the template DNA. The primers are short single-stranded DNA molecules (~20 bases long), one that is complementary to a portion of the 5'-3' strand, and another that is complementary to a portion of the 3'-5' strand of the template. These primers anneal to the separated template strands and serve as starting points for DNA Taq replication by DNA polymerase.
Taq DNA polymerase extends the annealed primers by "reading" the template strand and synthesizing the complementary sequence. In this way, Taq polymerase replicates the two template DNA strands. This polymerase has been isolated from a heat-stable bacterium (Thermus Aquaticus) that in nature lives within the steam vents in Yellowstone National Park. For this reason the enzymes within these bacteria have evolved to withstand high temperatures (94°C) and can be used in the PCR reaction.
PCR Step by Step
PCR amplification includes three main steps, a denaturation step, an annealing step, and an extension step (summarized in the figure below).
1) Denaturation: In denaturation the reaction mixture is heated to 94°C for 1 minute, which results in the melting or separation of the double-stranded DNA template into two single stranded molecules.
2) Amplification: In PCR amplification, DNA templates must be separated before the polymerase can generate a new copy. The high temperature required to melt the DNA strands normally would destroy the activity of most enzymes, but because Taq polymerase was isolated from bacteria that thrive in the high temperatures of hot springs, it remains active.
3) Annealing: During the annealing step, the oligonucleotide primers "anneal to" or find their complementary sequences on the two single-stranded template strands of DNA. In these annealed positions, they can act as primers for Taq DNA polymerase. They are called primers because they prime the synthesis of a new strand by providing a short sequence of double-stranded DNA for Taq polymerase to extend from and build a new complementary strand. Binding of the primers to their template sequences is also highly dependent on temperature.
During the extension step, the job of Taq DNA polymerase is to add nucleotides (A, T, G, and C) one at a time to the primer to create a complementary copy of the DNA template.

During polymerization the reaction temperature is 72°C, the temperature that produces optimal Taq polymerase activity. The three steps of denaturation, extension, and annealing form one "cycle" of PCR. A complete PCR amplification undergoes 40 cycles.
The entire 40-cycle reaction is carried out in a test tube placed into a thermal cycler. The thermal cycler contains an aluminum block that holds the samples and can be rapidly heated and cooled across broad temperature differences. The rapid heating and cooling of this thermal block is known as temperature cycling or thermal cycling.
PCR Temperature Cycle = Denaturation Step (94°C) + Annealing Step (60°C) + Extension Step (72°C)
DNA Fingerprinting
Variable Number of Tandem Repeats (VNTR)
The evolutionary principle of variation within a population is a cornerstone in biology. This variation results from subtle differences in the DNA sequence in individuals of a given species. One origin of variation results when duplication of a small sequence of nucleotides occurs during DNA replication. This results in a tandem repeat of the original sequence. If this mistake occurs again in another round of replication, then three copies of a sequence will be in tandem, as outlined below:
Individual 1: GTACTCCAATCATGTACCATGAC
Individual 2: GTACTCCAATCATCATGTACCATGAC
Individual 3: GTACTCCAATCATCATCATGTACCATGAC
Here single strands of DNA from the same locus and three different individuals are shown. 'CAT' is repeated once, twice or three times resulting in alleles of different lengths.
There are two standard methods for DNA fingerprinting:
- PCR of DNA containing VNTRs.
- Southern blotting (using RFLPs).
PCR allows the amplification of a single copy of DNA into millions of copies. However this technique requires that the DNA of interest already be of a known sequence in order to design primers that will specifically hybridize to the target DNA. One such region that is used in several countries for forensic analysis of DNA samples is the D1S80 locus. The D1S80 locus is located on the distal portion of the short arm of chromosome 1 and contains a variable number of tandem repeats.

D1S80 Example Tandem Repeats
Electrophoresis
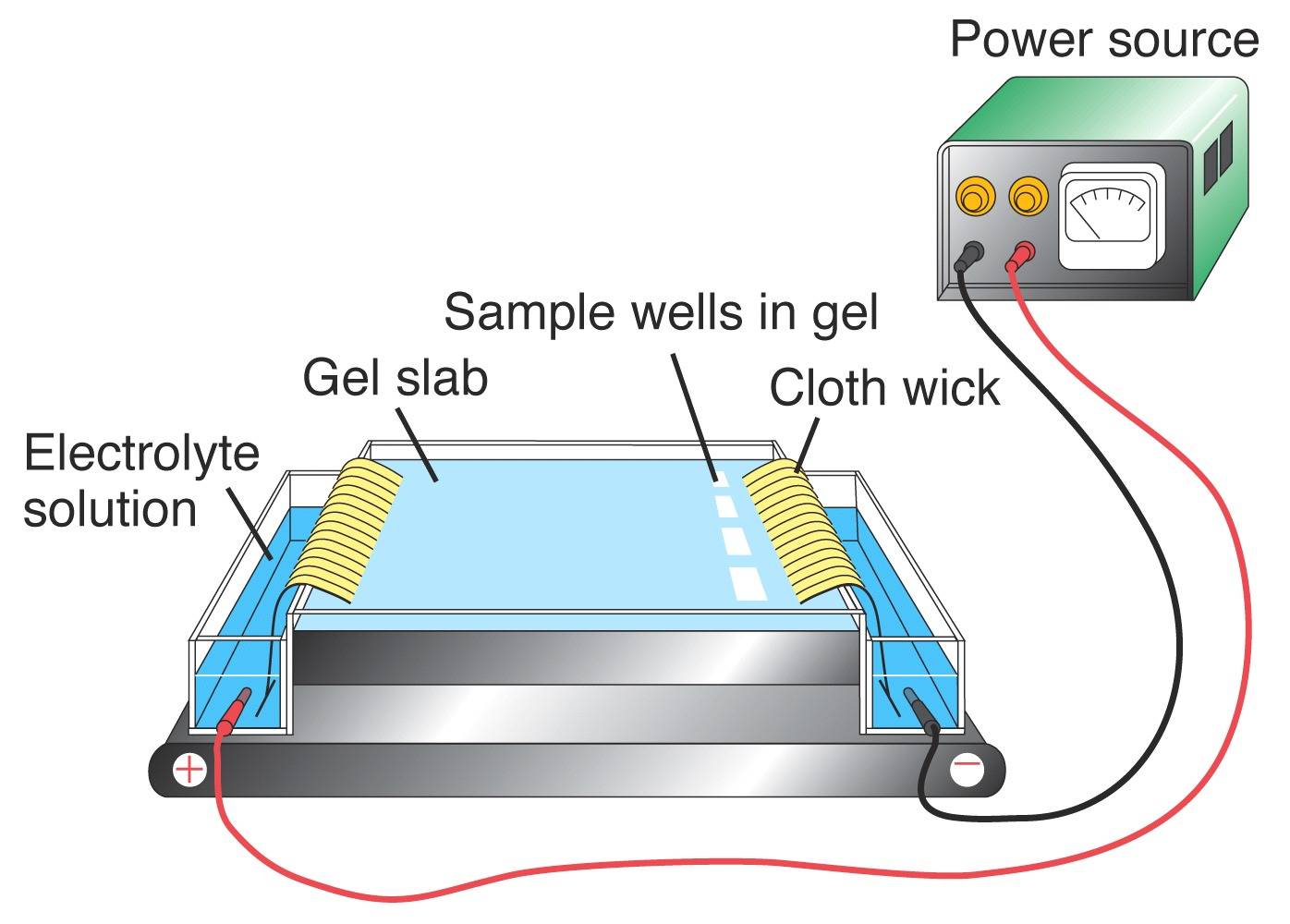
Using PCR, portions of DNA that are known to contain VNTRs can be amplified. The resultant product is then viewed to obtain the individuals DNA genotype. In order to visualize the DNA profile from a given locus, the DNA fragments need to be separated according to their sizes using gel electrophoresis. An electric current is applied to an agarose gel matrix. Since DNA is negatively charged due to the phosphate groups, the fragments will move towards the positive pole. DNA molecules of different sizes migrate through the gel at different speeds, with the smaller fragments moving faster that the larger ones. Once the DNA is stained in the gel, it can be visualized to reveal the DNA profile.

(Left) Fictitious agarose gel. Lanes 1, 2 and 3 represent bands using the data from the individuals listed above. Lane 4 represents DNA from a crime scene. (Right) Typical agarose gel apparatus.
Ethidium bromide (EtBr) is a large, flat basic molecule that resembles a DNA base pair.

Ethidium Bromide
Because of its chemical structure, it can intercalate (or insert) into a DNA strand. Ethidium bromide is commonly used in molecular biology laboratories to stain electrophoresis gels. The compound forms fluorescent complexes with nucleic acids and these can be viewed under UV light. EtBr is added to the warmed agarose prior to pouring the gel to give a final concentration of 0.5ug/ml. EtBr is a mutagen and must be handled with extreme caution using gloved hands.
Sequencing - Background
RFLP (often pronounced "rif lip", as if it were a word) is a method used by molecular biologists to follow a particular sequence of DNA as it is passed on to other cells. RFLPs can be used in many different settings to accomplish different objectives. RFLPs can be used in paternity cases or criminal cases to determine the source of a DNA sample. RFLPs can also be used determine the disease status of an individual. .
Each organism inherits its DNA from its parents. Since DNA is replicated with each generation, any given sequence can be passed on to the next generation. An RFLP is a sequence of DNA that has a restriction site on each end with a "target" sequence in between. A target sequence is any segment of DNA that can bind to a probe by forming complementary base pairs. A probe is a sequence of single-stranded DNA that has been tagged with radioactivity or an enzyme so that the probe can be detected. When a probe base pairs to its target, the investigator can detect this binding and know where the target sequence is since the probe is detectable. RFLP produces a series of bands when a Southern blot is performed with a particular combination of restriction enzyme and probe sequence.
For example, let's follow a particular RFLP that is defined by the restriction enzyme EcoR I and the target sequence of 20 bases GCATGCATGCATGCATGCAT. EcoR I binds to its recognition sequence GAATTC and cuts the double-stranded DNA as shown:

In the segment of DNA shown below, you can see the elements of an RFLP; a target sequence flanked by a pair of restriction sites. When this segment of DNA is cut by EcoR I, three restriction fragments are produced, but only one contains the target sequence which can be bound by the complementary probe sequence (purple).

Let's look at two people and the segments of DNA they carry that contain this RFLP (for clarity, we will only see one of the two stands of DNA). Since Jack and Jill are both diploid (double stranded DNA) organisms, they have two copies of this RFLP. When we examine one copy from Jack and one copy from Jill, we see that they are identical:
- Jack 1: -GAATTC---(8.2 kb)---GCATGCATGCATGCATGCAT---(4.2 kb)---GAATTC-
- Jill 1: -GAATTC---(8.2 kb)---GCATGCATGCATGCATGCAT---(4.2 kb)---GAATTC-
When we examine their second copies of this RFLP, we see that they are not identical. Jack 2 lacks an EcoR I restriction site that Jill has 1.2 kb upstream of the target sequence (difference in italics).
- Jack 2: -GAATTC--(1.8 kb)-CCCTTT--(1.2 kb)--GCATGCATGCATGCATGCAT--(1.3 kb)-GAATTC-
- Jill 2: -GAATTC--(1.8 kb)-GAATTC--(1.2 kb)--GCATGCATGCATGCATGCAT--(1.3 kb)-GAATTC-
Therefore, when Jack and Jill have their DNA subject to RFLP analysis, they will have one band in common and one band that does not match the other's in molecular weight:

Southern Blot Method
This is a brief overview of how a Southern blot (more formally called an DNA blot) is performed and what type of data you can obtain form one. The outcome of a Southern Blot is what most people think of when they refer to a "DNA Fingerprint".

Southern blots allow investigators to determine the molecular weight of a restriction fragment and to measure relative amounts in different samples.
Procedure:
- DNA (genomic or other source) is digested with a restriction enzyme and separated by gel electrophoresis, usually an agarose gel. Because there are so many different restriction fragments on the gel, it usually appears as a smear rather than discrete bands. The DNA is denatured into single strands by incubation with NaOH.
- The DNA is transferred to a membrane which is a sheet of special blotting paper. The DNA fragments retain the same pattern of separation they had on the gel.
- The blot is incubated with many copies of a probe which is single-stranded DNA. This probe will form base pairs with its complementary DNA sequence and bind to form a double-stranded DNA molecule. The probe cannot be seen but it is either radioactive or has an enzyme bound to it (e.g. alkaline phosphatase or horseradish peroxidase).
- The location of the probe is revealed by incubating it with a colorless substrate that the attached enzyme converts to a colored product that can be seen or gives off light which will expose X-ray film. If the probe was labeled with radioactivity, it can expose X-ray film directly.
Below is an example of a real Southern blot used to detect the presence of a gene that was transformed into a mixed cell population. In this Southern blot, it is easy to determine which cells incorporated the gene and which ones did not.

The figure on the left shows a photograph of a 0.7% agarose gel that has 14 different samples loaded on it (plus molecular weight marker in the far right lane and a glowing ruler used for analysis of the results). Each sample of DNA has been digested with the same restriction enzyme (EcoRI). Notice that the DNA does not appear as a series of discrete bands but rather as a smear. The figure on the right is a copy of the X-ray film and reveals which strains contain the target DNA and which ones do not.
A Laser scan of the gel produces a genomic fingerprint of the type below:
